微生物学講座 研究
研究
主に口腔疾患の原因菌が保有する病原因子に関して、分子生物学的手法で研究を行っていますが、これに限らず、広く微生物学・口腔細菌学の諸問題に関心を持っており、興味深いと思われる新しいことがらにも随時取り組んでいます。

(1)歯周病関連細菌の定着機構の解明
歯周病の発症や進行に関連する主要な細菌(ポルフィロモナス・ジンジバリス、トレポネーマ・デンティコーラ、タンネレラ・フォーシシア等)の定着機構に関する研究を行っている。これらの細菌は、他の細菌と凝集したり、宿主組織に付着したりして、歯周組織に定着する。これらの細菌の菌体表面分子(線毛や外膜タンパク質等)に注目し、それらの構造、形成機序および機能解析を、主に分子生物学的手法を用いて検討している。
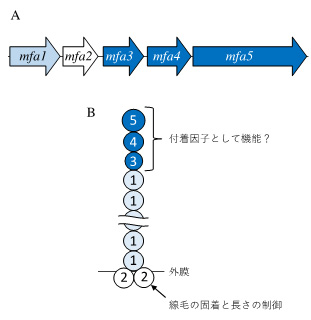
図は、ポルフィロモナス・ジンジバリスのMfa1線毛の構成成分をコードする遺伝子(A)と推定される構造(B)を示す。主要成分Mfa1が線毛のシャフトを構成し、その先端にMfa3、Mfa4およびMfa5が局在する。アンカーとして機能するMfa2は線毛基部に局在しダイマーあるいはトライマーを形成する。先端因子が宿主組織への付着において重要な役割を演じていることが考えられる。
詳細は以下のホームページを参照
Hasegawa Y:Elucidation of biogenesis mechanism and structure of fimbria from periodontal pathogen porphyromonas gingivalis.Impact,7:59-61.2019,
https://www.ingentaconnect.com/content/sil/impact/2019/00002019/00000007/art00021
Hasegawa Y:Periodontal disease:Structure of Mfa 1 fimbriae.OpenAccessGovernment,Oct:62-63.2019.
(2)歯周病関連細菌の短鎖脂肪酸の合成機構の解明とその応用に関する研究
歯周病細菌が産生する酪酸等の短鎖脂肪酸は、重度の歯周病患者の歯周ポケット内においては、健常人の10倍程度の高濃度で検出される。また、ヒト培養細胞にアポトーシスを起こす、バイオフィルム形成を促進する、感染ウイルスを活性化する等の作用が示唆されている。本研究課題では、①歯周病関連細菌における短鎖脂肪酸の産生機構の解明、②生体への為害作用やバイオフィルム増加の分子レベルでの解明、③短鎖脂肪酸の歯周病の診断ツールとしての開発、④酪酸産生酵素のリード化合物を利用した短鎖脂肪酸産生の抑制薬物の開発を目指す。
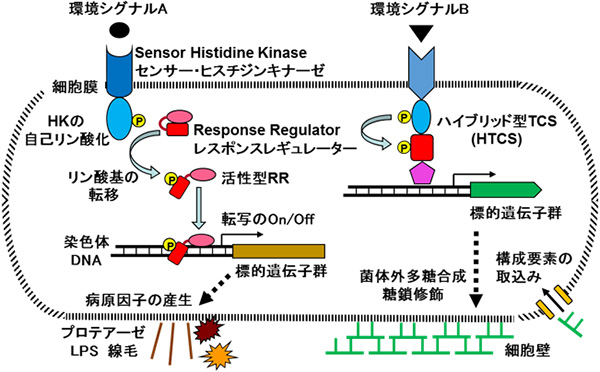
(3)歯周病関連細菌の病原性を調節する2成分制御系に関する研究
細菌は生息環境の変化に適応するために独特のシグナル伝達機構を持っている。最も典型的なものは2成分制御系(TCS)で、物理化学的刺激や分子などの環境シグナルを感知するセンサー・ヒスチジンキナーゼと、それからのシグナルに呼応して関連遺伝子の発現調節を行うレスポンスレギュレーターから成る。主要な歯周病関連細菌のポルフィロモナス・ジンジバリスやタンネレラ・フォーサイシアにも各々10前後のTCSが見出され、病原性の発現に特化した系の存在も明らかになってきた。これらのTCS1つ1つが受容する環境シグナル及びその制御下にある標的遺伝子群の同定を目指し、主に分子遺伝学的手法を用いて研究している。
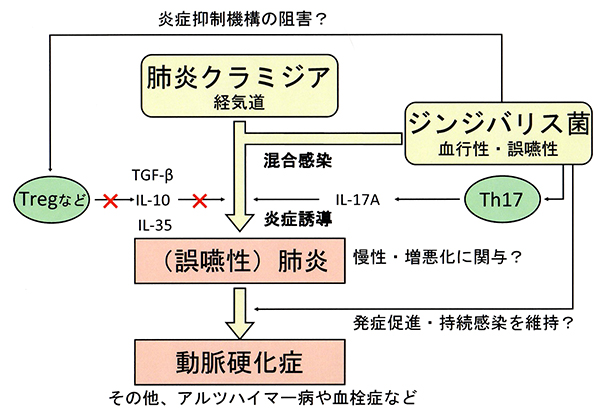
(4)歯周病原菌と肺炎クラミジア菌の混合感染が肺炎や動脈硬化症の誘発・増悪化におよぼす影響の研究
肺炎クラミジア菌は歯周炎に罹患した歯肉縁下プラークでも歯周病原菌と共に高頻度で検出される。歯周炎は肺炎や動脈硬化症との関連が疫学的に証明されている。歯周炎や肺炎、動脈硬化症の発症ならびに増悪化にヘルパーT[Th]l7細胞が、またその抑制には制御性T[Treg]細胞やIL-35産生細胞等が関与している可能性が示唆されているが、その誘導や制御に関しての詳細な機序は不明である。本研究では、肺炎クラミジア菌と歯周病酸をマウスに混合感染させ、その炎症過程における肺炎の増悪化ならびにその後の動脈硬化症への誘発・促進機序を明らかにすると共に、肺炎の起炎菌として長期細胞内で生存する肺炎クラミジアの宿主側の防御機構からのエスケープ機構を明らかにし、肺炎クラミジアに対するより効果的な治療法の確立を目指す。
主な研究業績
2021年
- Nagano K and Hasegawa Y: Periodoutal pathogens. Methods and protocols. Springer Nature, 2021.
- Nishiyama, SI, Hasegawa, Y, Nagano, K: Site-directed and random mutagenesis in Porphyromonas gingivalis: Construction of fimbriae-related-gene mutant. Methods Mol Biol, 2210:3-14, 2021.
- Nagano, K, Hasegawa, Y: Construction of a gene-deletion mutant in Tannerella forsythia. Methods Mol Biol, 2210:25-31, 2021.
- Hasegawa, Y, Nagano, K, Murakami, Y, Lamont, R.J: Purification of native Mfal fimbriae from Porphyromonas gingivalis. Methods Mol Biol, 2210:75-86, 2021.
- Murakami, Y, Nagano, K, Hasegawa, Y: Separation of glycosylated OmpA-Like proteins from Porphyromonas gingivalis and Tannerella forsythia. Methods Mol Biol, 2210:143-155, 2021.
- Yoshida, Y: Analysis of the butyrate-producing pathway in Porphyromonas gingivalis. Methods Mol Biol, 2210:167-172. 2021.
- Hirose, M, Yoshida, Y, Horii, K, Hasegawa, Y, Shibuya, Y: Efficacy of antimicrobial photodynamic therapy with Rose Bengal and blue light against cariogenic bacteria. Arch Oral Biol, 122: 105024, 2021.
- Fujinami W, Nishikawa K, Ozawa S, Hasegawa Y, Takebe J: Correlation between the relative Fujinal abundance of oral bacteria and Candida albicans in denture and dental plaques. J Oral Biosci, 63:175-183, 2021.
- Sakae, K, Nagano, K, Furuhashi, M, Hasegawa, Y: Diversity analysis of genes encoding Mfal fimbrial components in Porphyromonas gingivalis strains. PLoS ONE, 16(7):e 0255111, 2021.
- Hasegawa, Y, Nagano, K: Porphyromonas gingivalis FimA and Mfal fimbriae: Current insights on localization, function, biogenesis, and genotype. Jpn Dent Sci Rev,57:190-200, 2021.
- Okabe, T, Kamiya, Y, Kikuchi, T, Goto, H, Umemura, M, Suzuki, Y, Sugita, Y, Naiki, Y, Hasegawa, Y, Hayashi, J.-1, Kawamura, S, Sawada, N, Takayanagi, Y, Fujimura, T, Higuchi, N, Mitani, A: Porphyromonas gingivalis components/secretions synergistically enhance pneumonia caused by streptococcus pneumoniae in mice. Int J Mol Sci, 22: 12704, 2021.
- Okumura S, Konishi Y, Narukawa M, Sugiura Y, Yoshimoto S, Arai Y, Sato S, Yoshida Y, Tsuji S, Uemura K, Wakita M, Matsudaira T, Matsumoto T, Kawamoto S, Takahashi A, Itatani Y, Miki H, Takamatsu M, Obama K, Takeuchi K, Suematsu M, Ohtani N, Fukunaga Y, Ueno M, Sakai Y, Nagayama S, Hara E. Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat Comm, 12(1):5674, 2021.
- Hirose M, Yoshida Y, Horii K, Hasegawa Y, Shibuya Y. Effect of antimicrobial photodynamic therapy on periodontopathogenic bacteria: Evaluation of Rose Bengal and riboflavin as photosensitizers (in English), The Jpn J Oral Infect Dis, 28(2): 69-77, 2021.
2020年
- Takayanagi Y, Kikuchi T, Hasegawa Y, Naiki Y, Goto H, Okada K, Okabe I. Kamiya Y, Suzuki Y, Sawada N, Okabe T, Suzuki Y, Kondo S, Ohno T, Hayashi JI, Mitani A: Porphyromonas gingiva!is Mfa 1 induces chemokine and cell adhesion molecules in mouse gingival fibroblasts via Toll-like receptors. J Clin Med. 10;9(12):4004. 2020.
2019年
- Yoshida Y,SatoM,Nonaka T,Hasegawa Y,Kezuka Y:Characterization of the phosphotransacetylase-acetate kinase pathway for ATP production in Porphyromonas gingivalis.J.Oral Microbiol,11:1588086.2019.
- Sato-Boku A,Nagano K,Hasegawa Y,Kamimura Y,Sento Y,So M,Kako E,Okuda M,Tachi N,Ito H,Adachi Y,Sobue K:Comparison o fdisinfection effct between benzalkonium chloride and povidone iodine in nasotracheal intubation:a randomized trial.BMC Anesthesiol,19(1):168,2019.
- 西川 清,長谷川義明:デンチャープラーク微生物叢の群衆解析と義歯ケア・日本歯科医師会雑誌,72(5):17-26,2019.
2018年
- Hall M, Hasegawa Y, Yoshimura F, Persson K. Structural and functional characterization of shaft, anchor, and tip proteins of the Mfa1 fimbria from the periodontal pathogen Porphyromonas gingivalis. Sci Rep. 29;8(1):1793, 2018.
- Kezuka Y, Ishida T, Yoshida Y, Nonaka T. Structural insights into the catalytic mechanism of cysteine (hydroxyl) lyase from the hydrogen sulfide-producing oral pathogen, Fusobacterium nucleatum. Biochem J. 16;475(4):733-748, 2018.
- Lee JY, Miller DP, Wu L, Casella CR, Hasegawa Y, Lamont RJ: Maturation of the Mfa1 fimbriae in the oral pathogen Porphyromonas gingivalis. Front Cell Infect Microbiol.9;8:137. 2018.
- Sato W, Yoshida Y, Komasa S, Hasegawa Y, Okazaki J:Antimicrobial effect of titanium hydroxyapatite in denture base resin. App Sci, 8(6): 963, 2018.
- Konno H, Yoshida Y, Nagano K, Takebe J, Hasegawa Y:Biological and biochemical roles of two distinct cyclic dimeric adenosine 3’,5’-monophosphate-associated phosphodiesterases in Streptococcus mutans. Front Microbiol, 9: 2347, 2018.
- Kondo Y, Sato K, Nagano K, Nishiguchi M, Hoshino T, Fujiwara T, Nakayama K: Involvement of PorK, a component of the type IX secretion system, in Prevotella melaninogenica pathogenicity Microbiol Immunol 62: 554-566, 2018.
- Nagano K, Hasegawa Y, Iijima Y, Kikuchi T, Mitani A: Distribution of Porphyromonas gingivalis fimA and mfa1 fimbrial genotypes in subgingival plaques PeerJ 6:e5581, 2018.
- 長谷川義明:歯周病原細菌Porphyromonas gingivalisのもつMfa1線毛の新しい構造モデルと機能.日本口腔感染症学会雑誌,25:3-7, 2018.
2017年
- Ohno T, Yamamoto G, Hayashi JI, Nishida E, Goto H, Sasaki Y, Kikuchi T, Fukuda M, Hasegawa Y, Mogi M, Mitani A. Angiopoietin-like protein 2 regulates Porphyromonas gingivalis lipopolysaccharide-induced inflammatory response in human gingival epithelial cells. PLoS One. 21;12(9):e0184825, 2017.
- Moriguchi K, Hasegawa Y, Higuchi N, Murakami Y, Yoshimura F, Nakata K, Honda M. Energy dispersive spectroscopy-scanning transmission electron microscope observations of free radical production in human polymorphonuclear leukocytes phagocytosing non-opsonized Tannerella forsythia. Microsc Res Tech. 80(6):555-562, 2017.
- Okada K, Fujimura T, Kikuchi T, Aino M, Kamiya Y, Izawa A, Iwamura Y, Goto H, Okabe I, Miyake E, Hasegawa Y, Mogi M, Mitani A. Effect of interleukin (IL)-35 on IL-17 expression and production by human CD4+ T cells. PeerJ. 15;5:e2999, 2017.
- Nagano K, Hasegawa Y, Yoshida Y, and Yoshimura F. Novel fimbrilin PGN_1808 in Porphyromonas gingivalis. PLoS One. 12(3): e0173541, 2017.
- Ruscitto A, Honma K, Veeramachineni V, Nishikawa K, Stafford G, Sharma A. Regulation and molecular basis of environmental muropeptide uptake and utilization in fastidious oral anaerobe Tannerella forsythia. Front. Microbiol, 8: 648, 2017
- Nagano K, Hasegawa Y, Yoshida Y, Yoshimura F. Comparative analysis of motility and other properties of Treponema denticola strains. Microb Pathog. 102:82-88, 2017.
2016年
- Kloppsteck P, Hall M, Hasegawa Y, Persson K: Structure of the fimbrial protein Mfa4 from Porphyromonas gingivalis in its precursor form: implications for a donor-strand complementation mechanism. Sci Rep, 14; 6: 22945, 2016.
- Yoshida Y, Sato M, Kezuka Y, Hasegawa Y, Nagano K, Takebe J, Yoshimura F. Acyl-CoA reductase producing succinate semialdehyde from succinyl-CoA in the butyrate production of Porphyromonas gingivalis. Arch Biochim Biophys, 596: 138-148, 2016.
- Hasegawa Y, Iijima Y, Persson K, Nagano K, Yoshida Y, Lamont RJ, Kikuchi T, Mitani A, Yoshimura F. Role of Mfa5 in expression of Mfa1 fimbriae in Porphyromonas gingivalis. J Dent Res, 95(11): 1291-1297, 2016.
- Mahanonda R, Champaiboon C, Subbalekha K, Sa-Ard-Iam N, Rattanathammatada W, Thawanaphong S, Rerkyen P, Yoshimura F, Nagano K, Lang NP, Pichyangkul S: Human memory B cells in healthy gingiva, gingivitis, and periodontitis. J Immunol, 197(3): 715-725, 2016.
- Tomino M, Nagano K, Hayashi T, Kuroki K, Kawai T: Antimicrobial efficacy of gutta-percha supplemented with cetylpyridinium chloride. J Oral Sci, 58(2): 277-282, 2016.
- Sato M, Yoshida Y, Nagano K, Hasegawa Y, Takebe J, Yoshimura F: Three CoA transferases involved in the production of short chain fatty acids in Porphyromonas gingivalis. Front Microbiol. 7: 1146, 2016.
- Nishida E, Aino M, Kobayashi S, Okada K, Ohno T, Kikuchi T, Hayashi J, Yamamoto G, Hasegawa Y, Mitani A: Serum amyloid A promotes E-selectin expression via toll-like receptor 2 in human aortic endothelial cells. Mediators Inflamm, 2016: 7150509, 2016.
- Horie T, Inomata M, Into T, Hasegawa Y, Kitai N, Yoshimura F, Murakami Y: Identification of OmpA-like protein of Tannerella forsythia as an O-linked glycoprotein and its binding capability to lectins. PLoS One, 11(10): e0163974, 2016.
- Izumigawa M, Hasegawa Y, Ikai R, Horie T, Inomata M, Into T, Kitai N, Yoshimura F, Murakami Y: Separation of novel phosphoproteins of Porphyromonas gingivalis using phosphate-affinity chromatography. Microbiol Immunol, 60(10): 702-707, 2016.
2015年
- Hutcherson JA, Bagaitkar J, Nagano K, Yoshimura F, Wang H, Scott DA. Porphyromonas gingivalis RagB is a proinflammatory signal transducer and activator of transcription 4 agonist. Mol Oral Microbiol. 30 (3):242-252, 2015.
- Cueno ME, Nagano K, Imai K, Tamura M, Yoshimura F, Ochiai K. Ab initio modeling approach towards establishing the structure and docking orientation of the Porphyromonas gingivalis FimA. J Mol Graph Model. 55:65-71, 2015.
- Nagano K, Hasegawa Y, Yoshida Y, Yoshimura F: A major fimbrilin variant of Mfa1 fimbriae in Porphyromonas gingivalis. J Dent Res. 94 (8): 1143-1148, 2015.
- Konno H, Yoshida Y, Nagano K, Hasegawa Y, Nakamura Y, Tanaka Y, Yoshimura F: Quantification of bacterial biofilms on different thermoplastic denture base resins using immunoblotting with enhanced chemiluminescence. Aichi Gakuin Dent Sci. 28:11-18, 2015.
- Ikai R, Hasegawa Y, Izumigawa M, Nagano K, Yoshida Y, Kitai N, Lamont RJ, Yoshimura F, Murakami Y. Mfa4, an accessory protein of Mfa1 fimbriae, modulates fimbrial biogenesis, cell auto-aggregation, and biofilm formation in Porphyromonas gingivalis. PLoS One. 5 (10): e0139454, 2015.
- Goto T, Nagano K, Hirakawa H, Tanaka K, Yoshimura F. Draft genome sequence of Porphyromonas gingivalis strain Ando expressing a 53-kilodalton-type fimbrilin variant of Mfa1 fimbriae. Genome Announc. 3 (6). pii: e01292-15, 2015.
- Yoshida Y, Sato M, Nagano K, Hasegawa Y, Okamoto T, Yoshimura F. Production of 4-hydroxybutyrate from succinate semialdehyde in butyrate biosynthesis in Porphyromonas gingivalis. Biochim Biophys Acta. 1850 (12):2582-2591, 2015.
2014年
- Abiko Y, Nagano K, Yoshida Y, Yoshimura F. Major membrane protein TDE2508 regulates adhesive potency in Treponema denticola. PLoS One, 9 (2): e89051, 2014.
- Yang J, Yoshida Y, Cisar JO. Genetic basis of coaggregation receptor polysaccharide biosynthesis in Streptococcus sanguinis and related species. Mol Oral Microbiol, 29 (1): 24-31, 2014.
- Hasegawa Y, Murakami Y. Porphyromonas gingivalis fimbriae: Recent developments describing the function and localization of mfa1 gene cluster proteins. J. Oral Biosci, 56 (3): 86-90, 2014.
- Yoshida Y, Yang J, Nagano K, Yoshimura F, Cisar JO. Cell surface coaggregation receptor polysaccharide of oral streptococci. J. Oral Biosci, 56 (4): 125-130, 2014.
- Yoshida Y, Konno H, Nagano K, Abiko Y, Nakamura Y, Tanaka Y, Yoshimura F. The influence of a glucosyltransferase, encoded by gtfP, on biofilm formation by Streptococcus sanguinis in a dual-species model. APMIS, 122 (10): 951-960, 2014.
- Narita Y, Sato K, Yukitake H, Shoji M, Nakane D, Nagano K, Yoshimura F, Naito M, Nakayama K. Lack of a surface layer in Tannerella forsythia mutants deficient in the type IX secretion system. Microbiology-SGM, 160 (Pt 10): 2295-2303, 2014.
- Murakami Y, Hasegawa Y, Nagano K, Yoshimura F. Characterization of wheat germ agglutinin lectin-reactive glycosylated OmpA-like proteins derived from Porphyromonas gingivalis. Infect. Immun, 82 (11), 4563-4571, 2014.
- Abiko Y, Nagano K, Yoshida Y, Yoshimura F. Characterization of Treponema denticola mutants defective in the major antigenic proteins, Msp and TmpC. PLoS One, 9 (11): e113565, 2014.
- Nagano K, Taguchi K, Tokoro S, Tatsuno I, Mori H. Adhesion of enterohemorrhagic Escherichia coli O157:H7 to the intestinal epithelia is essential for inducing secretory IgA antibody production in the intestine of mice. Biol Pharm Bull, 37 (3): 409-416, 2014.
2013年
- Nagano K, Abiko Y, Yoshida Y, Yoshimura F. Genetic and antigenic analyses of Porphyromonas gingivalis FimA fimbriae. Mol Oral Microbiol. 28 (5): 392-403, 2013.
- Kezuka Y, Yoshida Y, Nonaka T. Crystal structures of βC-S lyase from Streptococcus anginosus in complex with its reaction intermediates. Photon Factory Activity Rep. Part B. 30: 275, 2013.
- Yang J, Yoshida Y, Cisar JO. Genetic basis of coaggregation receptor polysaccharide biosynthesis in Streptococcus sanguinis and related species. Mol Oral Microbiol. 29 (1): 24-31, 2013.
- Hasegawa Y, Nagano K, Ikai R, Izumigawa M, Yoshida Y, Kitai N, Lamont RJ, Murakami Y, Yoshimura F. Localization and function of the accessary protein Mfa3 in Porphyromonas gingivalis Mfa1 fimbriae. Mol Oral Microbiol. 28 (6): 469-480, 2013.
- Hajishengallis G, McIntosh ML, Nishiyama S, Yoshimura F. Mechanism and Implications of CXCR4-Mediated Integrin Activation by Porphyromonas gingivalis. Mol. Oral Microbiol. Mol Oral Microbiol. 28 (4): 239-249, 2013.
- Shimotahira N, Oogai Y, Kawada-Matsuo M, Yamada S, Fukutsuji K, Nagano K, Yoshimura F, Noguchi K, Komatsuzawa H. The S-layer of Tannerella forsythia Contributes to Serum Resistance and Oral Bacterial Co-aggregation. Infect Immun. 81 (4): 1198-1206, 2013.
- 永野恵司. 歯周病関連細菌Porphyromonas gingivalisのFimA線毛に関する研究. 薬学雑誌. 133: 963-974, 2013.
- Nishikawa K, Tanaka Y. A simple mutagenesis using natural competence in Tannerella forsythia. J Microbiol Methods, 94(3): 378-380, 2013.
2012年
- Yano A, Kikuchi S, Takahashi T, Kohama K, Yoshida Y. Inhibitory effects of the phenolic fraction from the pomace of Vitis coignetiae on biofilm formation by Streptococcus mutans. Arch Oral Biol. 57 (6) :711-719, 2012.
- Virtuoso LP, Harden JL, Sotomayor P, Sigurdson WJ, Yoshimura F, Egilmez NK, Minev B, Kilinc MO. Characterization of iNOS (+) Neutrophil-like ring cell in tumor-bearing mice. J Transl Med. 10:152, 2012.
- Nagano K, Abiko Y, Yoshida Y, Yoshimura F. Porphyromonas gingivalis FimA fimbriae: Roles of the fim gene cluster in the fimbrial assembly and antigenic heterogeneity among fimA genotypes. J Oral Biosci. 54 (3): 160-163, 2012.
- Sugawara E, Nagano K, Nikaido H. Alternative folding pathways of the major porin OprF of Pseudomonas aeruginosa. FEBS J. 279 (6): 910-918, 2012.
- Kezuka Y, Abe N, Yoshida Y, Nonaka T. Purification, crystallization and preliminary X-ray analysis of two hydrogen sulfide-producing enzymes from Fusobacterium nucleatum. Acta Cryst. F. 68 (12): 1507-1510, 2012.
- Kezuka Y, Yoshida Y, Nonaka T. Structural insights into catalysis by βC-S lyase from Streptococcus anginosus. Proteins. 80: 2447-2458, 2012.
- de Toledo A, Nagata E, Yoshida Y, Oho T. Streptococcus oralis coaggregation receptor polysaccharides induce inflammatory responses in human aortic endothelial cells. Mol Oral Microbiol. 27 (4): 295-307, 2012.
- Kishi M, Hasegawa Y, Nagano K, Nakamura H, Murakami Y, Yoshimura F. Identification and characterization of novel glycoproteins involved in growth and biofilm formation by Porphyromonas gingivalis. Mol Oral Microbiol. 27 (6): 458-470, 2012.
- Nagano K, Hasegawa Y, Abiko Y, Yoshida Y, Murakami Y, Yoshimura F. Porphyromonas gingivalis FimA fimbriae: Fimbrial assembly by fimA alone in the fim gene cluster and differential antigenicity among fimA genotypes. PLoS One 7 (9): e43722, 2012.
- Komatsu T, Nagano K, Sugiura S, Hagiwara M, Tanigawa N, Abiko Y, Yoshimura F, Furuichi Y, Matsushita K. E-selectin mediates Porphyromonas gingivalis adherence to human endothelial cells. Infect Immun. 80 (7): 2570-2576, 2012.

